This year Ximbio's Anti-Cisplatin Modified DNA, clone CP9/19 was published in a particularly exciting Scientific Reports paper. The publication, from Professor Simon Reed's lab at the University of Cardiff, details a novel technique known as 3D-DIP-Chip, which can be used to measure the prevalence of genomic genotoxin-induced DNA damage.
The 3D-DIP-Chip method is a microarray-based method for the genome-wide analysis of DNA damage and repair characteristics. The protocol is summarised as follows:
(a) Proteins are crosslinked to chromatin (top) or DNA damage is induced (bottom). This DNA or chromatin is extracted, sonicated and split into two samples.
(b) IP is carried out on one sample to separate the chromatin bound factor of interest (top) or damaged DNA (bottom).
(c) Both samples are purified, amplified by PCR and differentially labelled with red or green fluorophores.
(d) The samples are allowed to hybridise to the microarray probes and the resulting intensity values from the scanned image are converted to numerical values.
(e) These values may be plotted and processed as required by the investigation.
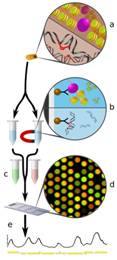
Cartoon representation of the 3D-DIP-Chip process. (Powell et al. 2015. Sci Rep. 5:7975.)
Clone CP9/19, which has been highly cited, recognises only the intra-strand cross-links formed by cisplatin between adjacent purines. It was developed by Dr Michael Tilby at the University of Newcastle. Michael spent his career researching the mechanism of action of DNA damage inducing anticancer drugs.
HAVE YOU REGISTERED?
Join the ever-growing global Ximbio community. Register and receive updates about new reagents, institutes and new features being added to the website.
